vanderPloeg2024_analysis
Source:vignettes/vanderPloeg2024_analysis.Rmd
vanderPloeg2024_analysis.RmdIntroduction
In this vignette a PARAFAC model is created for upper jaw lingual
data from the vanderPloeg2024 study. This is done by first
processing the count data. Subsequently, the appropriate number of
components are determined. Then the PARAFAC model is created and
visualized.
Processing the data cube
The data cube in vanderPloeg2024$upper_jaw_lingual$data
contains unprocessed counts. The function processDataCube()
performs the processing of these counts with the following steps:
- It performs feature selection based on the sparsityThreshold
setting. Sparsity is here defined as the fraction of samples where a
microbial abundance (ASV/OTU or otherwise) is zero. For
vanderPloeg2024we can take the RFgroups groups into account for feature selection. We do this by calculating the sparsity for each feature in each subject group and compare those against the sparsity threshold that we set. If a feature passes the threshold in either group, it is selected. - It performs a centered log-ratio transformation of each sample using
the
compositions::clr()function with a pseudo-count of one (on all features, prior to selection based on sparsity). - It centers and scales the three-way array. This is a complex subject, for which we refer to a paper by Rasmus Bro and Age Smilde. By centering across the subject mode, we make the subjects comparable to each other within each time point. Scaling within the feature mode avoids the PARAFAC model focusing on features with abnormally high variation.
The outcome of processing is a new version of the dataset called
processedPloeg. Please refer to the documentation of
processDataCube() for more information.
processedPloeg = processDataCube(vanderPloeg2024$upper_jaw_lingual, sparsityThreshold=0.50, considerGroups=TRUE, groupVariable="RFgroup", CLR=TRUE, centerMode=1, scaleMode=2)Determining the correct number of components
A critical aspect of PARAFAC modelling is to determine the correct
number of components. We have developed the functions
assessModelQuality() and
assessModelStability() for this purpose. First, we will
assess the model quality and specify the minimum and maximum number of
components to investigate and the number of randomly initialized models
to try for each number of components.
Note: this vignette reflects a minimum working example for analyzing
this dataset due to computational limitations in automatic vignette
rendering. Hence, we only look at 1-3 components with 5 random
initializations each. These settings are not ideal for real datasets.
Please refer to the documentation of assessModelQuality()
for more information.
# Setup
# For computational purposes we deviate from the default settings
minNumComponents = 1
maxNumComponents = 3
numRepetitions = 3 # number of randomly initialized models
numFolds = 5 # number of jack-knifed models
ctol = 1e-5
maxit = 200
numCores = 1
colourCols = c("RFgroup", "Phylum", "")
legendTitles = c("RF group", "Phylum", "")
xLabels = c("Subject index", "Feature index", "Time index")
legendColNums = c(3,5,0)
arrangeModes = c(TRUE, TRUE, FALSE)
continuousModes = c(FALSE,FALSE,TRUE)
# Assess the metrics to determine the correct number of components
qualityAssessment = assessModelQuality(processedPloeg$data, minNumComponents, maxNumComponents, numRepetitions, ctol=ctol, maxit=maxit, numCores=numCores)The overview plot showcases the number of iterations, the sum-of-squared error, the CORCONDIA and the variance explained for 1-3 components.
qualityAssessment$plots$overview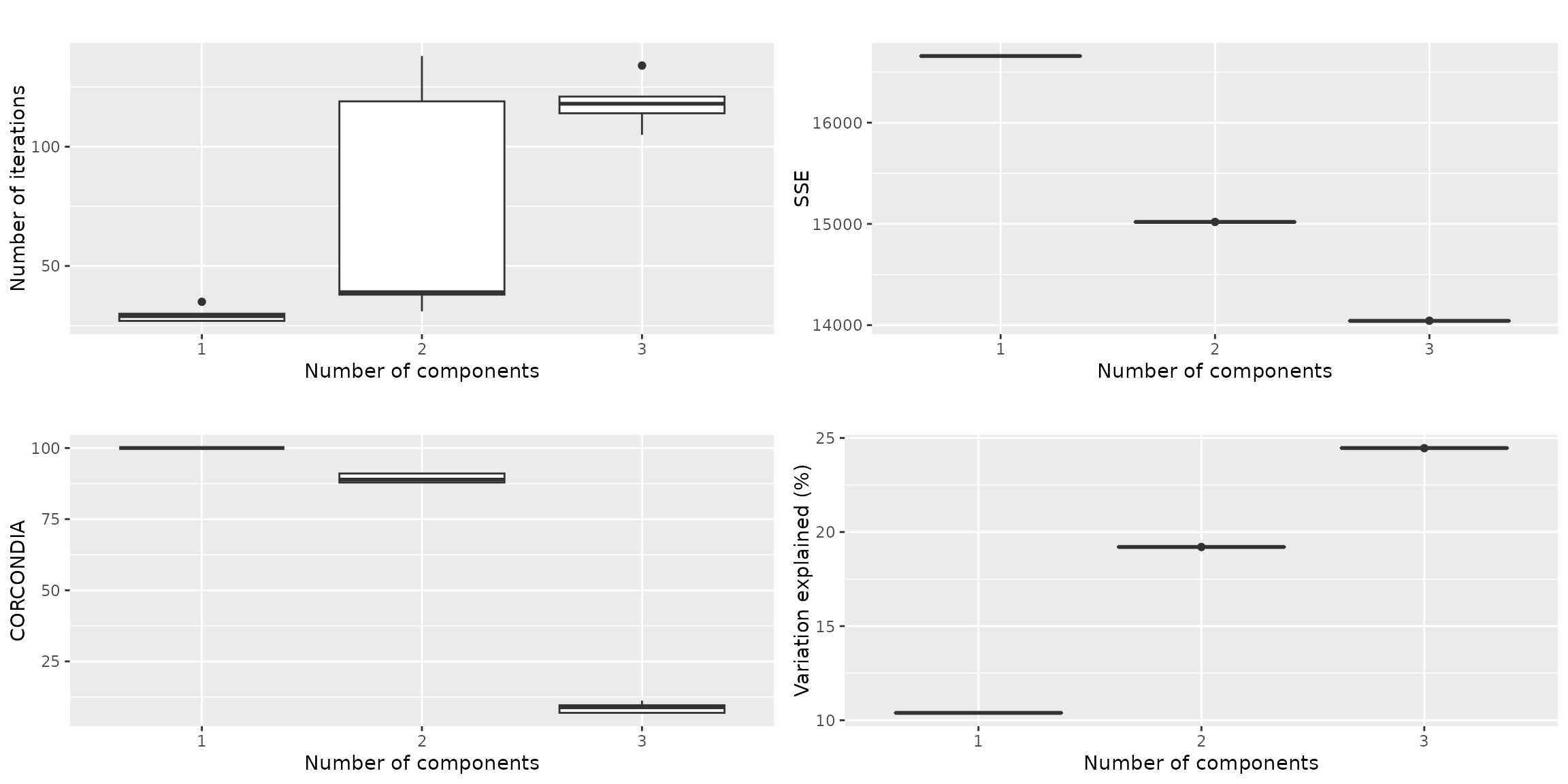 This seems a clear-cut case for a two-component model, as the
three-component models have a CORCONDIA near zero. The maximum amount of
variation we can describe is ~20%. We skip the stability assessment
since the appropriate number of components is clearly two.
This seems a clear-cut case for a two-component model, as the
three-component models have a CORCONDIA near zero. The maximum amount of
variation we can describe is ~20%. We skip the stability assessment
since the appropriate number of components is clearly two.
Model selection
We have decided that a two-component model is the most appropriate
for the vanderPloeg2024 dataset. We can now select one of
the random initializations from the assessModelQuality()
output as our final model. We’re going to select the random
initialization that corresponded the maximum amount of variation
explained for two components.
numComponents = 2
modelChoice = which(qualityAssessment$metrics$varExp[,numComponents] == max(qualityAssessment$metrics$varExp[,numComponents]))
finalModel = qualityAssessment$models[[numComponents]][[modelChoice]]Finally, we visualize the model using
plotPARAFACmodel().
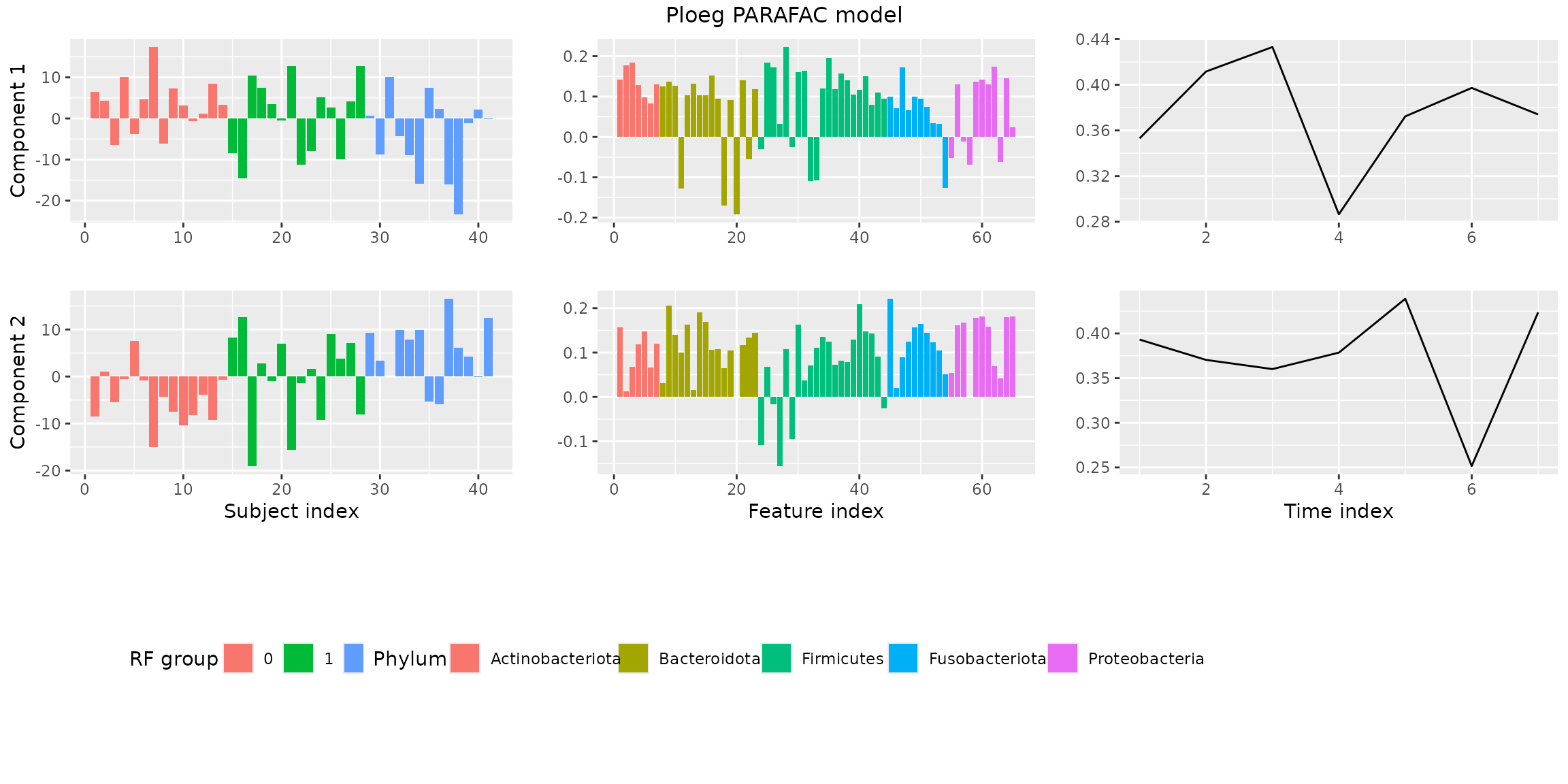
plotPARAFACmodel(finalModel$Fac, processedPloeg, 2, colourCols, legendTitles, xLabels, legendColNums, arrangeModes, continuousModes,
overallTitle = "vanderPloeg2024 PARAFAC model")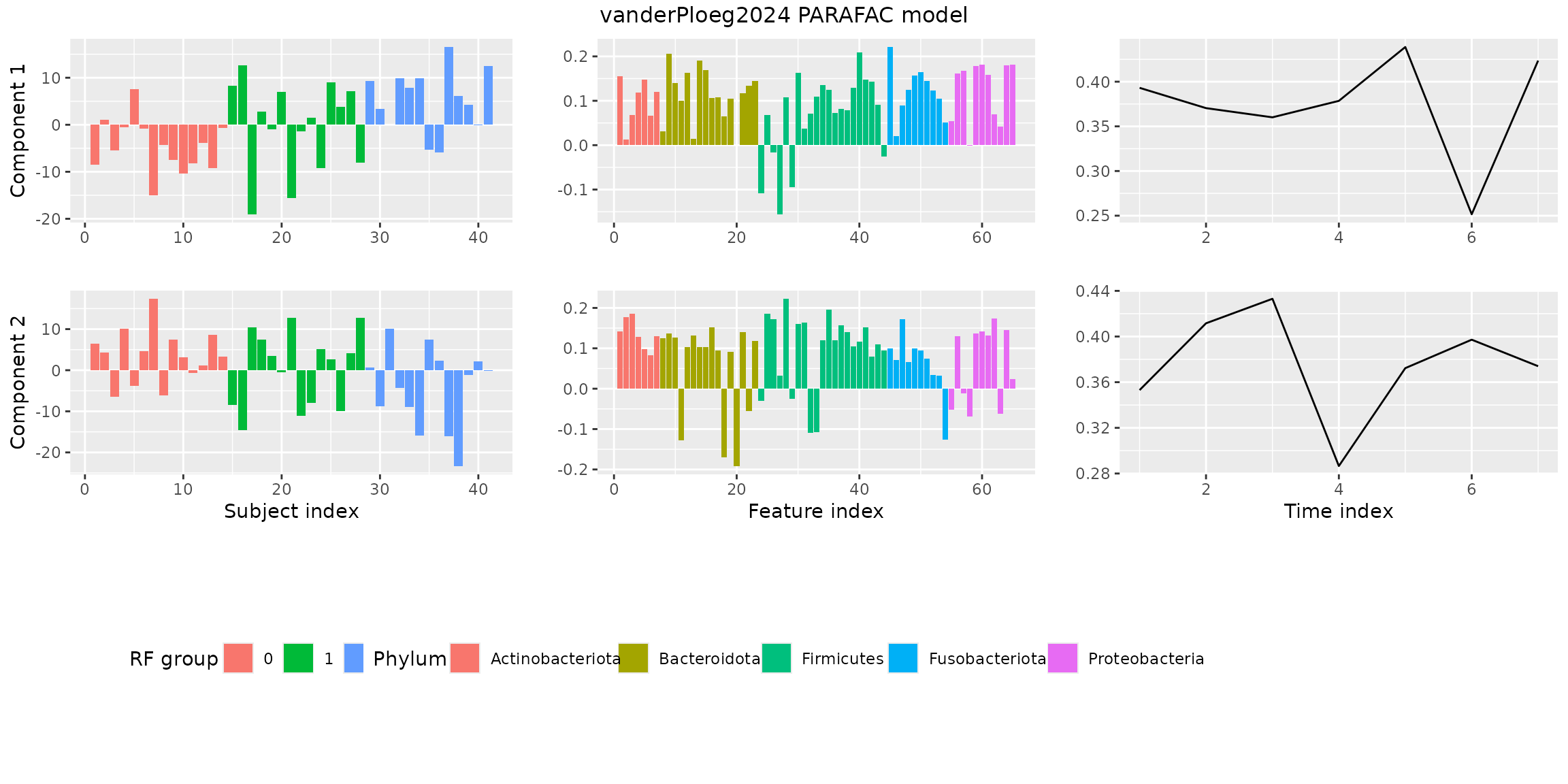
You will observe that the loadings for some modes in some components
are all negative. This is due to sign flipping: two modes having
negative loadings cancel out but describe the same thing as two positive
loadings. The flipLoadings() function automatically
performs this procedure and also sorts the components by how much
variation they describe.
finalModel = flipLoadings(finalModel, processedPloeg$data)
plotPARAFACmodel(finalModel$Fac, processedPloeg, 2, colourCols, legendTitles, xLabels, legendColNums, arrangeModes,
continuousModes = c(FALSE,FALSE,TRUE),
overallTitle = "Ploeg PARAFAC model")